| 主要代表性论文
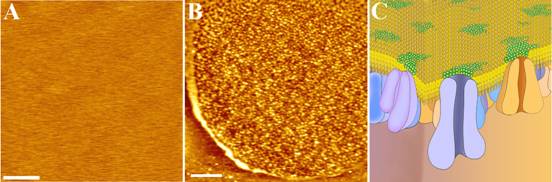
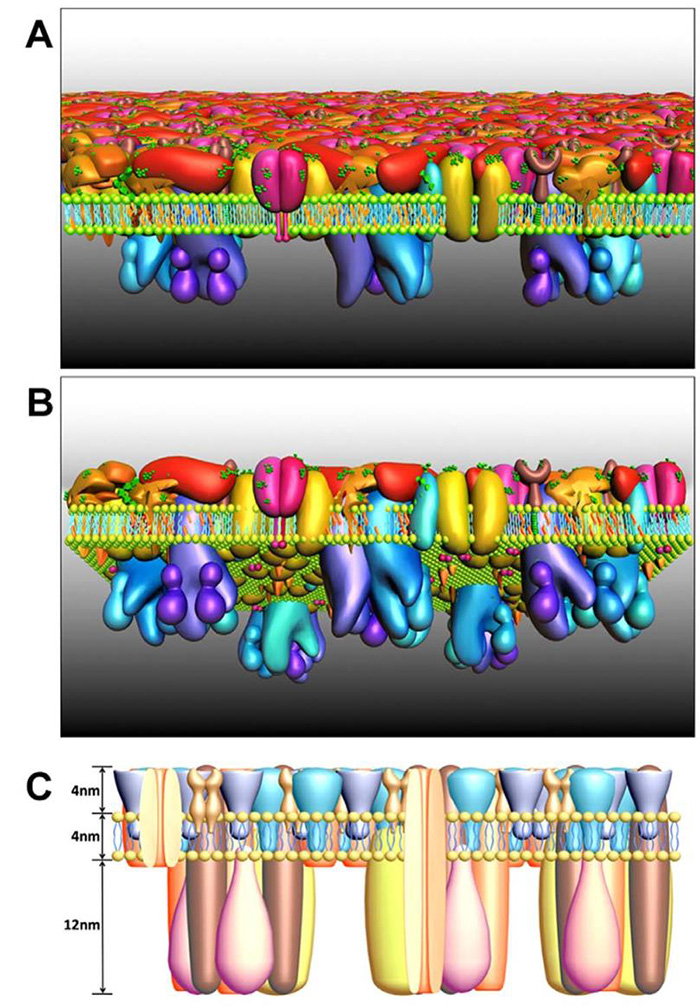
1. Qiuyan Yan, Yanting Lu, Lulu Zhou, Junling Chen, Haijiao Xu, Mingjun Cai, Yan Shi, Junguang Jiang, Wenyong Xiong*, Jing Gao*, Hongda Wang*, Mechanistic insights into GLUT1 activation and clustering revealed by super-resolution imaging, Proceedings of the National Academy of Sciences, 2018, 115(27): 7033-7038. IF: 9.504
2. Yangang Pan, Yuebin Zhang, Pianchou Gongpan, Qingrong Zhang, Siteng Huang, Bin Wang, BingqianXu, YupingShan*, Wenyong Xiong*, Guohui Li*, Hongda Wang*, Single glucose molecule transport process revealed by force tracing and molecular dynamics simulations,Nanoscale Horizons, 2018, 3(5): 517-524. IF: 9.391
3. Jing Gao#, Lingli He#, Yan Shi#, Mingjun Cai, Haijiao Xu, Junguang Jiang, Lei Zhang*, Hongda Wang*, Cell Contact and Pressure Control of Yap Localization and Clustering Revealed by Super-Resolution Imaging, Nanoscale, 2017, 9(43):16993-7003. IF: 7.233
4. Yangang Pan, Fuxian Zhang, Liuyang Zhang, Shuheng Liu, Mingjun Cai, Yuping Shan*, Xiaoqiao Wang*, Hanzhong Wang*, Hongda Wang*, The Process of Wrapping Virus Revealed by a Force Tracing Technique and Simulations. Advanced Science. 2017, 4(9):7. DIO: 10.1002/advs.201600489. IF: 12.441
5. Junling Chen, Tianzhou Liu, Jing Gao, Lan Gao, Lulu Zhou, Mingjun Cai, Yan Shi, Wenyong Xiong, Junguang Jiang, Ti Tong*, Hongda Wang*, Variation in carbohydrates between cancer and normal cell membranes revealed by super-resolution fluorescence imaging, Advanced Science, 2016, 3(12):9. DIO: 10.1002/advs.201600270. IF: 12.441
6. Junling Chen, Jing Gao, Mingjun Cai, Haijiao Xu, Junguang Jiang, Zhiyuan Tian, Hongda Wang*, Mechanistic insights into the distribution of carbohydrate clusters on cell membranes revealed by dSTORM imaging, Nanoscale, 2016, 8(28):13611-13619. IF: 7.233
7. Yuping Shan, Hongda Wang*, The structure and function of cell membranes examined by atomic force microscopy and single-molecule force spectroscopy, Chemical Society Reviews, 2015, 44(11):3617-3638. IF: 40.182
8. Bohua Ding, Yongmei Tian, Yangang Pan, Yuping Shan, Mingjun Cai, Haijiao Xu, Yingchun Sun*, Hongda Wang*, Recording the dynamic endocytosis of single gold nanoparticles by AFM-based force tracing, Nanoscale, 2015, 7(17):7545-7549. IF: 7.233
9. Jing Gao, Ye Wang, Mingjun Cai, Yangang Pan, Haijiao Xu, Junguang Jiang Hongbin Ji*, Hongda, Wang*, Mechanistic insights into EGFR membrane clustering revealed by super-resolution imaging, Nanoscale, 2015, 7(6):2511-2519. IF: 7.233
10. Junling Chen, Jing Gao, Jiazhen Wu, Min Zhang, mingjun Cai, Haijiao Xu, Junguang Jiang, Zhiyuan Tian, Hongda, Wang*, Revealing the carbohydrate pattern on the cell surface by super-resolution imaging, Nanoscale, 2015, 7(8), 3373-3380. IF: 7.233
11. Yangang Pan, Shaowen Wang, Yuping Shan, Dinglin Zhang, Jing Gao, Min Zhang, Shuheng Liu, Mingjun Cai, Haijiao Xu, Guohui Li*, Qiwei Qin*, Hongda Wang*, Ultrafast tracking of a single live virion during the invagination of a cell membrane, Small, 2015, 11(23):2782-2788. IF: 9.598
12. Ye Wang#, Jing Gao#, Xingdong Guo#, Ti Tong, Xiaoshan Shi, Lunyi Li, Miao Qi, Yajie Wang, Mingjun Cai, Junguang Jiang, Chenqi Xu*, Hongbin Ji*, Hongda Wang*, Regulation of EGFR nanocluster formation by ionic protein-lipid interaction, Cell Research, 2014, 24(8):959-976. IF: 15.393
13. Yangang Pan, Feng Wang, Yanhou Liu, Junguang Jiang, Yong-Guang Yang*, Hongda Wang*, Studying the mechanism of CD47-SIRP alpha interactions on red blood cells by single molecule force spectroscopy, Nanoscale, 2014, 6(17):9951-9954. IF: 7.233
14. Heng Liu, Hui Yang, Xian Hao, Haijiao Xu, Debao Xiao*, Hongda Wang*, Zhiyuan Tian*, Development of polymeric nanoprobes with improved lifetime dynamic range and stability for intracellular oxygen sensing, Small, 2013, 9(15):2639-2648. IF: 9.598
15. Weidong Zhao, Mingjun Cai, Haijiao Xu, Junguang Jiang, Hongda Wang*, Single-Molecule Force Spectroscopy study of the interactions between lectins and carbohydrates on cancer and normal cells, Nanoscale, 2013, 5(8), 3226-3229. IF: 7.233
16. Weidong Zhao, Shuheng Liu, Mingjun Cai, Haijiao Xu, Junguang Jiang, Hongda Wang*, Detection of carbohydrates on the surface of cancer and normal cells with topography and recognition imaging, Chemical Communications, 2013, 49(29): 2980-2982. IF: 6.29
17. Mingjun Cai#, Weidong Zhao#, Xin Shang, Junguang Jiang, Hongbin Ji, Zhiyong Tang*, Hongda Wang*, Direct evidence of lipid rafts by in-situ atomic force microscopy, Small, 2012, 8(8):1243-1250. IF: 9.598
18. Yuping Shan, Jinfeng Huang, Juanjuan Tan, Gui Gao, Shuheng Liu, Hongda Wang*, Yuxin Chen*, The study of single anticancer peptides interacting with HeLa cell membranes by single molecule force spectroscopy, Nanoscale, 2012, 4(4):1283-1286. IF: 7.233
19. Xian Hao, Xin Shang, Jiazhen Wu, Yuping Shan, Mingjun Cai, Junguang Jiang, Zhong Huang*, Zhiyong Tang*, Hongda Wang*, Single particle tracking of hepatitis-B-virus-like vesicles entry into cells, Small, 2011, 7(9):1212-1218. IF: 9.598
20. Yuping Shan, Xian Hao, Xin Shang, Mingjun Cai, Junguang Jiang, Zhiyong Tang*, Hongda Wang*,Recording force events of single quantum-dot endocytosis, Chemical Communications, 2011, 47(12):3377-3379. IF: 6.29
21. Hongda Wang*, Xian Hao, Yuping Shan, Junguang Jiang, Mingjun Cai, Xin Shang, Preparation of cell membranes for high resolution imaging by AFM, Ultramicroscopy, 2010, 110(4):305-312. IF: 2.929
22. Junguang Jiang, Xian Hao, Mingjun Cai, Yuping Shan, Xin Shang, Zhiyong Tang, Hongda Wang*, Localization of Na+-K+ ATPases in Quasi-Native Cell Membranes, Nano Letters, 2009, 9(12):4489-4493. IF: 12.08
23. Hongda Wang*, Yamini Dalal*, Stuart Lindsay, Steven Henikoff, Single-epitope recognition imaging of native chromatin, Epigenetics & Chromatin, 2008, 1:10, doi:10.1186/1756-8935-1-10. IF: 5.351
24. Hongda Wang, Linda Obenauer-Kutner, Mei Lin, Yunping Huang, Michael Grace, Stuart Lindsay*, Imaging Glycosylation, Journal of the American Chemical Society, 2008, 130(26):8154-8155. IF: 14.357
25. Yamini Dalal, Hongda Wang, Stuart Lindsay, Steven Henikoff*, Tetrameric Structure of Centromeric Nucleosomes in Interphase Drosophila Cells, PLoS Biology, 2007, 5(8):798-1809. IF: 9.163
26. Cordula Stroh#, Hongda Wang#, Ralph Bash, Brian Ashcroft, Jeremy Nelson, Hermann Gruber, Dennis Lohr, Stuart Lindsay*, Peter Hinterdorfer*, Single-molecule recognition imaging microscopy. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(34):12503-12507. IF: 9.504
27. Hongda Wang, Ralph Bash, Jaya. G. Yodh, Gorden Hager, Stuart Lindsay*, Dennis Lohr, Using Atomic Force Microscopy to Study Nucleosome Remodeling on Individual Nucleosomal Arrays in Situ. Biophysical Journal 2004, 87(3):1964-1971. IF: 3.495
28. Hongda Wang, Ralph Bash, Jaya G. Yodh, Gorden L. Hager, Dennis Lohr, Stuart M. Lindsay*, Glutaraldehyde Modified Mica: A New Surface for Atomic Force Microscopy of Chromatin. Biophysical Journal, 2002, 83(6):3619-3625. IF: 3.495 |